SEQUOIA AI Revolutionizes Cancer Diagnosis with Gene Expression Predictions
Stanford Medicine researchers have introduced SEQUOIA, an AI-powered tool that predicts the activity of thousands of genes in tumor cells using routine biopsy images. This breakthrough could significantly reduce the time and cost associated with cancer diagnostics and treatment planning, traditionally reliant on expensive genomic tests.
Revolutionizing Cancer Diagnostics
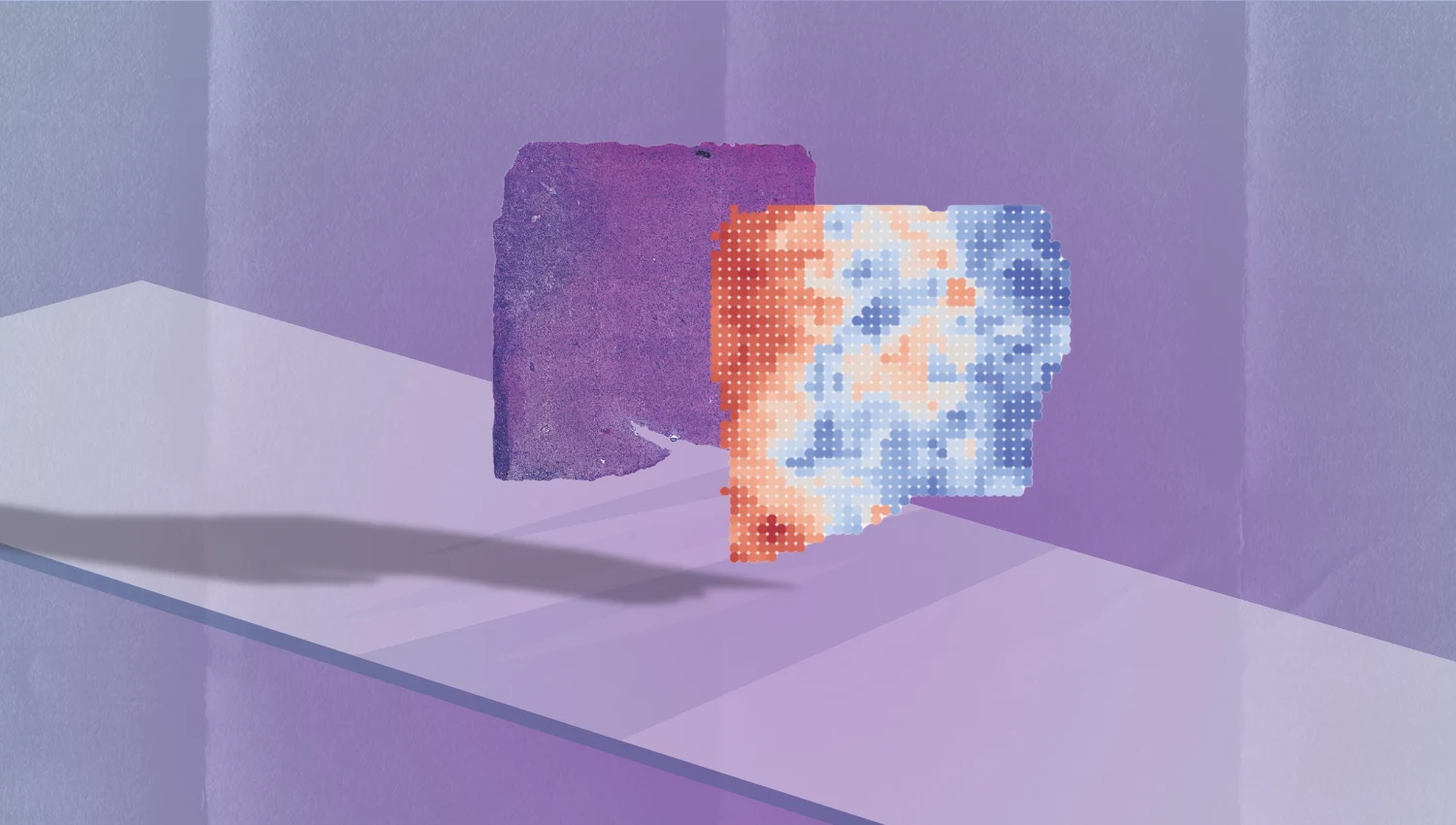
Traditionally, identifying the genetic drivers of cancer requires sequencing RNA from tumor biopsies—a process that is costly and time-intensive. SEQUOIA sidesteps this by analyzing the visual patterns of hematoxylin and eosin-stained biopsy slides. By doing so, it identifies gene expression patterns invisible to the human eye, enabling quicker clinical decision-making.
"This software could save the healthcare system thousands of dollars while speeding up treatment decisions," said Olivier Gevaert, PhD, the study's senior author.
How SEQUOIA Works
SEQUOIA was developed using a dataset of over 7,500 tumor samples spanning 16 cancer types. The AI model predicts the activation of over 15,000 genes based on biopsy images. In breast cancer, SEQUOIA accurately replicated results from FDA-approved genomic tests like MammaPrint, predicting risk scores from stained slides alone.
For some cancers, SEQUOIA achieved an 80% correlation with actual gene expression data. Additionally, it provides a visual genetic map of tumors, offering insights into genetic variations across different tumor regions.
Clinical Potential and Future Applications
While SEQUOIA isn't yet ready for clinical use, it represents a leap forward in oncology. The AI model is being refined for broader applications across all cancer types, potentially reducing dependency on expensive genetic tests.
As AI technologies like SEQUOIA evolve, they promise a future of faster, more accessible, and cost-effective cancer care.
Reference
For more details, visit Stanford Medicine.