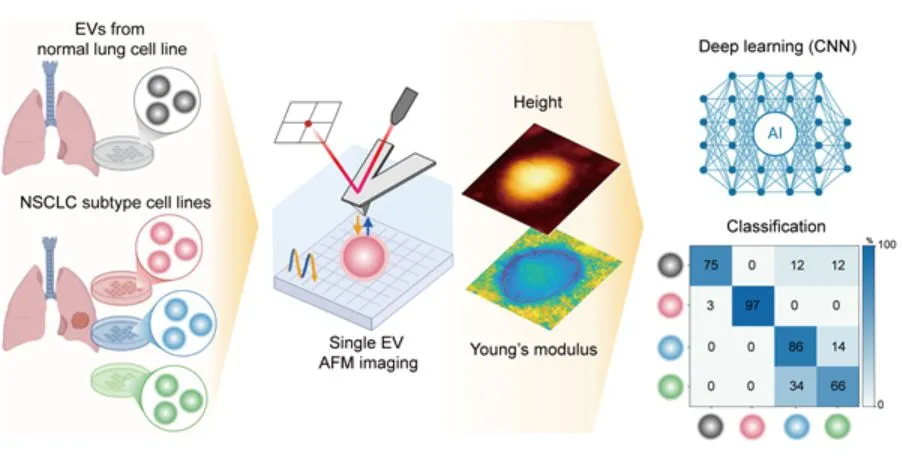
DGIST researchers developed a deep learning model that classifies lung cancer exosomes based on physical properties measured by atomic force microscopy.
Key Details
- 1DGIST team used AFM to measure nanomechanical properties (stiffness, height-to-radius) of exosomes from NSCLC cell lines with different genetic mutations.
- 2AI model (DenseNet-121) classified exosomes by origin, achieving 96% accuracy and AUC of 0.92.
- 3Exosome stiffness reflected KRAS and EGFR mutations in their respective lung cancer cell lines.
- 4The method enables high-precision, label-free, liquid biopsy-based lung cancer diagnosis.
- 5Study published July 8, 2025, in Analytical Chemistry.
Why It Matters

Source
EurekAlert
Related News

Deep Learning AI Outperforms Clinic Prognostics for Colorectal Cancer Recurrence
A new deep learning model using histopathology images identifies recurrence risk in stage II colorectal cancer more effectively than standard clinical predictors.

AI Reveals Key Health System Levers for Cancer Outcomes Globally
AI-based analysis identifies the most impactful policy and resource factors for improving cancer survival across 185 countries.

Dual-Branch Graph Attention Network Predicts ECT Success in Teen Depression
Researchers developed a dual-branch graph attention network that uses structural and functional MRI data to accurately predict individual responses to electroconvulsive therapy in adolescents with major depressive disorder.